New & Noteworthy
Give a Gift / Support SGD
January 31, 2025
Giving to SGD just got easier! We now accept donations by credit card with this form: give.stanford.edu.
Select ‘Other Stanford Designation’ under ‘Direct your gift’ & in the ‘Other’ box, add: Saccharomyces Genome Database – Account: GHJKO, Genetics: WAZC. Thanks for your support!
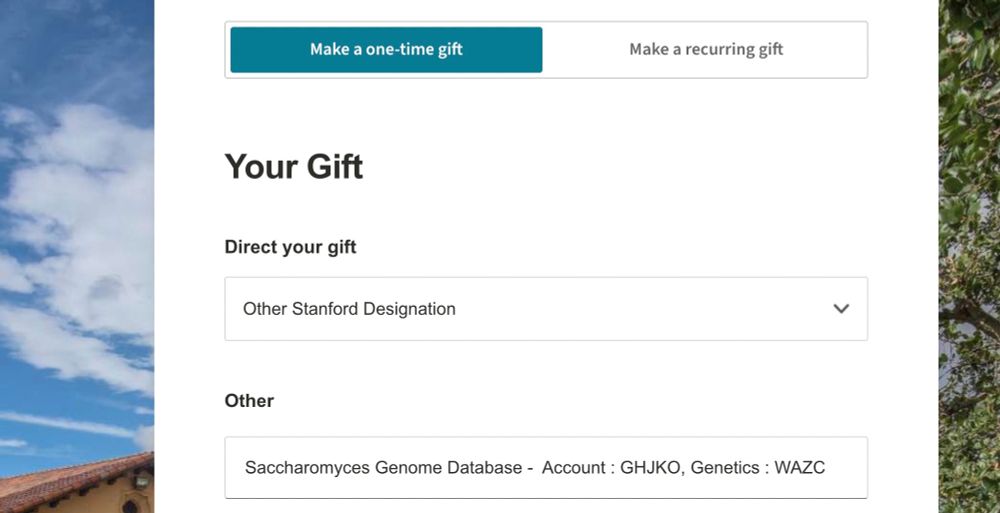
Your generous gift to SGD enables us to continue providing essential information for your research and teaching efforts. Donations are now critical for our work to continue.
To contribute via check, please make checks payable to Stanford University, and include a note stating that “these funds should be used to support the Saccharomyces Genome Database project in the Department of Genetics, Stanford University. Account : GHJKO, Genetics : WAZC.”
Thank you for your support!
Kindly send by mail to:
Development Services
PO Box 20466
Stanford, CA 94309
CONTACT US:
sgd-helpdesk@lists.stanford.edu
Categories: Announcements
AlphaFold protein structures now on SGD protein pages
November 18, 2024
We are thrilled to announce that we have now integrated AlphaFold protein structures into our protein pages! This cutting-edge addition provides detailed, high-accuracy 3D models of protein structures, offering invaluable insights into protein function and interactions. Researchers can now explore these comprehensive structural predictions directly within SGD, facilitating advanced studies in molecular biology and bioinformatics. Dive into the new AlphaFold protein structures and elevate your research with this powerful tool!

AlphaFold, developed by DeepMind, is an AI program that accurately predicts protein structures from amino acid sequences, enabling visualization of protein conformations. The predicted structures can be accessed through the Protein Data Bank (PDB) and AlphaFold Protein Structure Database.
Thanks to Kim Rutherford and Val Wood of Pombase for tips about adding AlphaFold structures to SGD.
Categories: Announcements
YeastMine shutting down July 15
July 01, 2024
Due to ongoing cuts to our funding, SGD can no longer continue to provide the YeastMine data warehouse resource. It is with heavy hearts that we discontinue this service.
Back in 2011, SGD implemented InterMine (http://www.InterMine.org), an open source data warehouse system with a sophisticated querying interface, to create YeastMine, a multifaceted search and retrieval environment that provided access to diverse data types. YeastMine served as a powerful search interface, a discovery tool, a curation aid, and a complex database presentation format.
YeastMine has served us all quite well. We are working to move the YeastMine data into AllianceMine, hosted by the Alliance of Genome Resources, of which SGD is a founding member.
To get started with AllianceMine, go to the Templates page, and filter by category = ‘YeastMine’.

Categories: Announcements
New user interface for YeastMine
April 04, 2024
Here at SGD we provide high-quality curated genomic, genetic, and molecular information on the genes and gene products of the budding yeast Saccharomyces cerevisiae. Twelve years ago, in order to accommodate the increasingly complex and diverse needs of researchers for searching and comparing data, SGD implemented InterMine, an open source data warehouse system with a sophisticated querying interface, to create YeastMine.
Today’s news is that we have updated YeastMine to use the new BlueGenes user interface from InterMine. The new interface provides the same functionality you’re already familiar with, wrapped in a new design offering a more interactive experience for exploring and analyzing your data. We are making this switch because AllianceMine at the Alliance of Genome Resources, of which SGD is a founding member, is using the updated interface as well.

YeastMine is a multifaceted search and retrieval environment that provides access to diverse data types. Searches can be initiated with a list of genes, a list of Gene Ontology terms, or lists of many other data types. The results from queries can be combined for further analysis and saved or downloaded in customizable file formats. Queries themselves can be customized by modifying predefined templates or by creating a new template to access a combination of specific data types.
User documentation for the new YeastMine interface is available from InterMine.
Categories: Announcements
Tags: user-interface
Changes to Saccharomyces cerevisiae GFF3 file
March 01, 2024
The saccharomyces_cerevisiae.gff contains sequence features of Saccharomyces cerevisiae and related information such as Locus descriptions and GO annotations. It is fully compatible with Generic Feature Format Version 3. It is updated weekly.
After November 2020, SGD updated the transcripts in the GFF file to reflect the experimentally determined transcripts (Pelechano et al. 2013, Ng et al. 2020), when possible. The longest transcripts were determined for two different growth media – galactose and dextrose. When available, experimentally determined transcripts for one or both conditions were added for a gene. When this data was absent, transcripts matching the start and stop coordinates of an open reading frame (ORF) were used.

Beginning in February 2024, SGD increased the start and stop coordinates of genes to encompass the start and stop coordinates of the longest experimentally determined transcripts, regardless of condition. This change was made in order to comply with JBrowse 2, a newer and more extensible genome browser, which requires that parent features in GFF files (genes) are larger than child features (mRNA, CDS, etc) (Diesh et al., 2023).

This is a standard format used by many groups. SGD uses the GFF file to load the reference tracks in SGD’s genome browser resource.
Categories: Announcements, Data updates
Tags: biology, blog, genetics, news, Saccharomyces cerevisiae
Search full-text with Textpresso: new papers added weekly
December 06, 2023
SGD’s instance of Textpresso has recently been updated! Each week, SGD biocurators triage new publications from PubMed to load the newest yeast papers into the database. Once they are in SGD, those papers get indexed and loaded into Textpresso – a tool for full-text mining and searching. This is the new part: Content updates in SGD’s Textpresso are now happening on a weekly basis, meaning you can search full text of the very latest yeast papers!

You already love Textpresso for searching full text and its other bells and whistles:
- Search results shown in the context of the full text – hits to query terms highlighted in situ
- Custom corpus creation – you can decide which papers to search
- Search using Boolean operators
- Search scope options for document or sentence
- Search location options can constrain to specific sections of papers

Textpresso can be accessed via the “Full-text Search” link under “Literature” in the purple toolbar that runs across the top of most SGD webpages. Now you can search full text of the very latest yeast papers each week!
Categories: Announcements
How do your papers get into SGD?
October 03, 2023
Members of the yeast community come to SGD to find the latest peer-reviewed budding yeast-specific scientific literature. You could go to Google or PubMed and find huge piles of literature…but then you’d have to slog through the lists and pages to find exactly what you’re looking for. Instead, you skip all that and come straight to SGD, knowing that SGD biocurators do all this vetting for you.
Each Friday night, we cast a wide net, scraping PubMed for any and all new papers, using keywords ‘yeast’ or ‘cerevisiae’. We do this broad search so that we don’t miss anything, but precisely because the search is so broad, we inevitably catch some papers that we end up throwing back. For the papers we keep each week, SGD biocurators “triage” them to attach genes, alleles, protein complexes, and biochemical pathways, and to identify papers that warrant further attention because they contain curatable phenotypes, functional annotations, regulatory relationships, post-translational modifications, disease associations, or large-scale datasets, etc.

SGD lists all the new papers on the ‘Literature Recently Added to SGD‘ page, with the newest papers added each day at the very top. This page includes all papers added to SGD within the past 30 days. In the past this page has just listed the papers. We recently made it more useful by adding the list of attached genes, alleles, protein complexes, and biochemical pathways (the ‘entities’) for each paper. Now you can search (i.e., ‘find in page’, ⌘+F, Ctrl+F) for your favorite gene(s) each week to see if any new papers have made it into SGD, so that you stay updated on research related to your area of study.
Access the ‘Literature Recently Added to SGD‘ page via the ‘New Yeast Papers‘ link in the Literature pull-down menu in the purple toolbar running across the top of most SGD webpages.

If you have a paper that should be in SGD but isn’t, please let us know so we can add it! Just shoot us an email, or use SGD’s Submit Data form (found in the Community pull-down menu in the purple toolbar running across the top of most SGD webpages).
Categories: Announcements, Website changes
GENETICS Knowledgebase and Database Resources
May 08, 2023
The May 2023 issue of GENETICS features the second annual collection of Model Organism Database articles. Scientists from Alliance of Genome Resources member groups SGD, RGD, ZFIN, Gene Ontology, and Xenbase have provided updates on recent activities and innovations. Be sure to browse the issue and get acquainted with these excellent Knowledgebase and Database Resource papers at GENETICS. Cover art by Vivid Biology.

Categories: Announcements
Tags: Saccharomyces cerevisiae, yeast
Apply Now for the 2023 Yeast Genetics and Genomics Course
March 10, 2023

For over 50 years, the legendary Yeast Genetics & Genomics course has been taught each summer at Cold Spring Harbor Laboratory. (OK, the name didn’t include “Genomics” in the beginning…). The list of people who have taken the course reads like a Who’s Who of yeast research, including Nobel laureates and many of today’s leading scientists.
The application deadline is April 1st, so don’t miss your chance!
Find all the details and application form at the CSHL Meetings & Courses site. This year’s instructors – Grant Brown, Maitreya Dunham, Soni Lacefield, and Greg Lang – have designed a course (July 25 – August 15) that provides a comprehensive education in all things yeast, from classical genetics through up-to-the-minute genomics. Students will perform and interpret experiments, learning about things like:
- Finding and Analyzing Yeast Information Using SGD
- Transformation & genome engineering
- Microscopy
- Manipulating yeast
- Dissecting tetrads
- Isolating mutants
- Working with essential genes
- Synthetic genetic arrays
- Fluctuation assays
- Whole genome sequencing & analysis
- Deep mutational scanning
Techniques have been summarized in the accompanying course manual, published by CSHL Press.

Scientists who aren’t part of large, well-known yeast labs are especially encouraged to apply – for example, professors and instructors who want to incorporate yeast into their undergraduate genetics classrooms; scientists who want to transition from mathematical, computational, or engineering disciplines into bench science; and researchers from small labs or institutions where it would otherwise be difficult to learn the fundamentals of yeast genetics and genomics. Significant stipends (in the 30-50% range of total fees) are available to individuals expressing a need for financial support and who are selected into the course.
Besides its scientific content, the fun and camaraderie at the course is also legendary. In between all the hard work there are late-night chats at the bar and swimming at the beach. There’s a fierce competition between students at the various CSHL courses in the Plate Race, which is a relay in which teams have to carry stacks of 40 Petri dishes (used, of course). There’s also typically a sailboat trip, a microscopy contest, and a mysterious “Dr. Evil” lab!
The Yeast Genetics & Genomics Course is loads of fun – don’t miss out!
Categories: Announcements, Conferences
Apply Now for the 2023 Fungal Pathogen Genomics Course
February 02, 2023

Fungal Pathogen Genomics is an exciting several day long course that provides experimental biologists working on fungal organisms with hands-on experience in genomic-scale data analysis. Through a collaborative teaching effort between the web-based fungal data mining resources FungiDB, EnsemblFungi, PomBase, SGD, CGD, MycoCosm, and JGI, students will learn how to utilize the unique tools provided by each database, develop testable hypotheses, and analyze various ‘omics’ datasets across multiple databases.
Daily activities will include individual and group training exercises, supplementary lectures on bioinformatics techniques and tools used by various databases, and presentations by distinguished guest speakers covering the following topics:
- Comparative genomics, gene trees, whole-genome alignment
- Identification of orthologs and orthology-based inference
- Gene pages and genome browsers
- RNA-Seq analysis and visualization in VEuPathDB Galaxy
- Variant calling analysis and Ensembl Variant Effect Predictor (VEP) tool
- Development of advanced biologically relevant queries using FungiDB ‘search strategies’ and mining integrated datasets (proteomics, transcriptomics, phenotypes, etc.)
- Genetic interactions, virulence genes, secondary metabolites
- Overview of ontology structure, evidence, available tools, slimming and enrichment
- Introduction to manual genome annotation and curation using Apollo, a web-based platform for structural and functional genome annotation (MycoCosm, Ensembl Fungi, FungiDB)
The application deadline for the Fungal Pathogen Genomics workshop to be held May 9-13, 2023 is February 16, 2023.
Don’t miss out – apply now!
Categories: Announcements, Conferences